For a full list of tools developed by our lab, please visit our
Github page.
For more detail, please refer to the publication .

For more detail, please refer to the publication .

DeepRepeat:
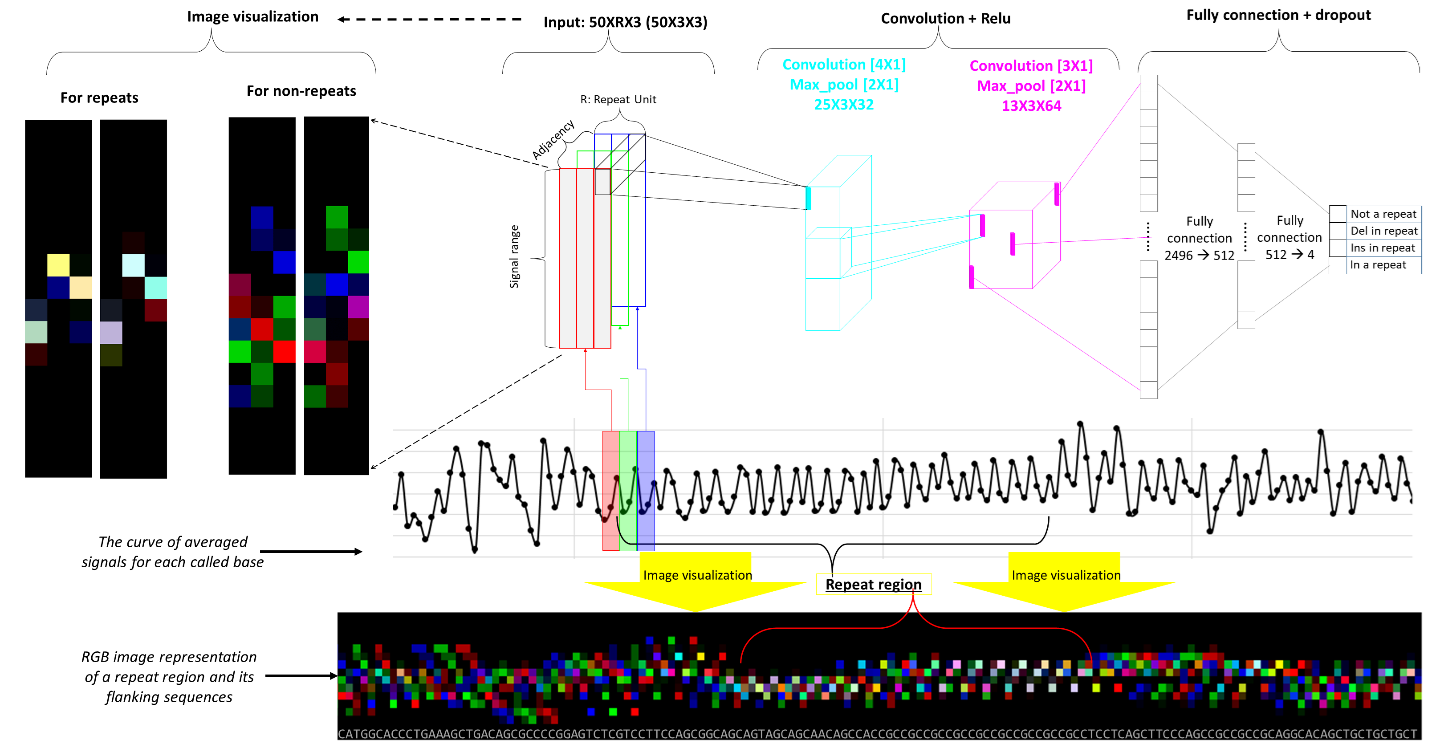
https://github.com/WGLab/DeepRepeat DeepRepeat is a deep learning estimation of short tandem repeats on Oxford Nanopore sequencing signals data. It takes the benefits of self-similarity of directly adjacent repeats and convert a repeat and its upstream and downstream repeats into RGB channels of a color image. And then, a deep convolutional neural network is used to learn repeat patterns followed by a summary by Gaussian mixture distribution for multiple reads.
DeepMod:
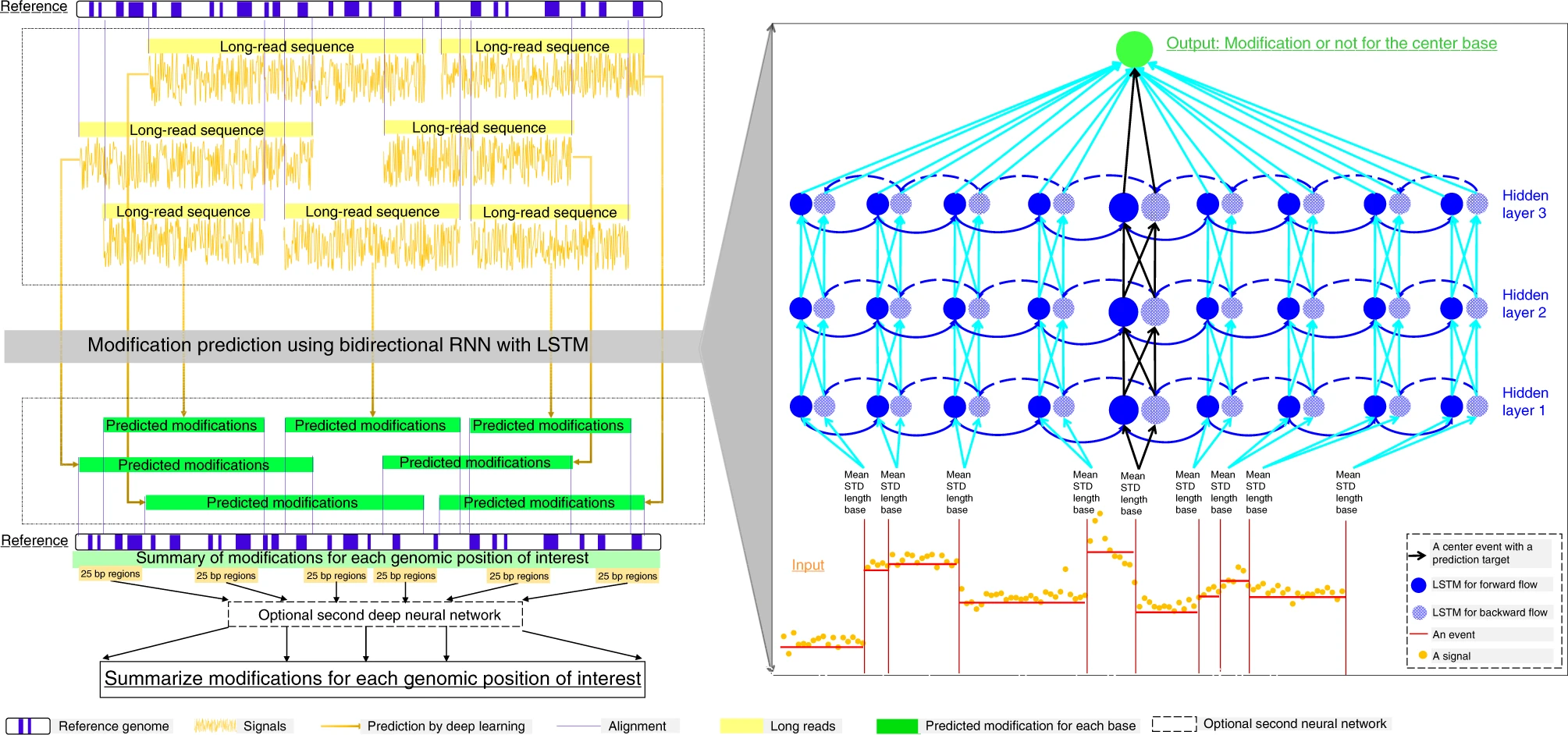
https://github.com/WGLab/DeepMod DeepMod is a deep-learning tool for genomic-scale, strand-sensitive and single-nucleotide based detection of DNA modifications. It is based on bidirectional recurrent neural network (RNN) with long short-term memory (LSTM) units.For more detail, please refer to the publication .

RepeatHMM:
https://github.com/WGLab/RepeatHMM RepeatHMM is a novel computational tool to detect any microsatellites (including trinucleotide repeats in trinucleotide repeat disorders (TRD)) from given long reads for a subject of interests. It is based on hidden Markov Model.For more detail, please refer to the publication .
